MOCAP data extraction and visualization
Motion Capture Technology
Short for MOCAP, this is used to record the movement of objects or, mainly people. The recorded data can be further utilized for other purpose in military, entertainment, sports, medical applications, also for creation and validation of computer vision and robotics.
Overview to CMU MOCAP Database
This is a famous motion dataset created by Carnegie Mellon University Graphics Lab.
It contains 2605 trials of motions in 6 categories and 23 subcategories by 111 human subjects. Humans wear a suit with 41 markers which can be captured with 12 Vicon infrared MX-40 cameras, each of which is capable of recording 120 Hz with images of 4 megapixel resolution.
The 3D data is provided in two way:
- Marker position, in .c3d files format.
- Skeleton movement, in a .vsk_.v pair or .asf_.amc pair, a .bvh format is also created and provided by other researchers.
Here we use the original .asf/.amc pair to extract the 3D data and to visualize.
.asf/.amc file format
.asf/.amc was created by a game company Acclaim which no longer exist.
.asf for skeleton
an example of asf file from 01 subject:
# AST/ASF file generated using VICON BodyLanguage
# -----------------------------------------------
:version 1.10 // Version of the skeleton definition
:name VICON // Arbitrary name for the skeleton
:units // Units definition
mass 1.0
length 0.45 // In this CMU dataset, all the data are multiplied by 0.45 in *INCHES*
angle deg // Euler angles are in degrees
:documentation // Arbitrary documentation
.ast/.asf automatically generated from VICON data using
VICON BodyBuilder and BodyLanguage model FoxedUp or BRILLIANT.MOD
:root // This is the root of the skeleton
order TX TY TZ RX RY RZ // The order of appearence of 6 degrees of freedom in AMC file
axis XYZ // Rotation order or the root
position 0 0 0 // Initial position/offset in World Coodinate System
orientation 0 0 0 // Initial orientation in WCS
:bonedata // Here starts the definition of each bone segment
begin
id 1 // Unique id for each bone
name lhipjoint // Unique name for each bone
direction 0.566809 -0.746272 0.349008
// Vector describing direction of the bone segment from
// parent to child in world coordinate system
length 2.40479 // Length of the bone segment
// The above direction and length of a segment determine
//the offset of the child from the parent
axis 0 0 0 XYZ // Rotation of local coordinate system for
//this bone relative to the world coordinate
//system. In .amc file the rotation angles
//for this bone for each time frame will be
//defined relative to this local coordinate
//system
end
begin
id 2
name lfemur
direction 0.34202 -0.939693 0
length 7.1578
axis 0 0 20 XYZ
dof rx ry rz // Not always 3 degrees of freedom
limits (-160.0 20.0) // Limits of each rotation
(-70.0 70.0)
(-60.0 70.0)
end
...
...
...
:hierarchy
// This part defines the hierarchy of each bone
// e.g. root is the parent to lhipjoint, rhipjoint and lowerback
begin
root lhipjoint rhipjoint lowerback
lhipjoint lfemur
lfemur ltibia
ltibia lfoot
lfoot ltoes
rhipjoint rfemur
rfemur rtibia
rtibia rfoot
rfoot rtoes
lowerback upperback
upperback thorax
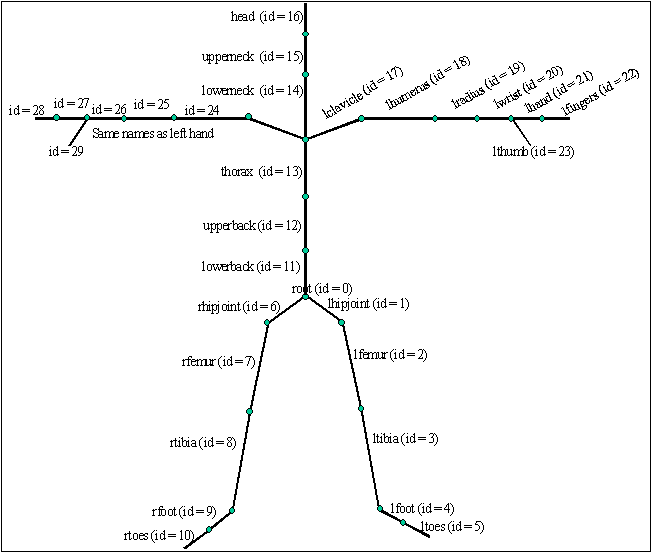
thorax lowerneck lclavicle rclavicle
lowerneck upperneck
upperneck head
lclavicle lhumerus
lhumerus lradius
lradius lwrist
lwrist lhand lthumb
lhand lfingers
rclavicle rhumerus
rhumerus rradius
rradius rwrist
rwrist rhand rthumb
rhand rfingers
end
Fig 1: Definition of the skeleton
Some notes:
- The WCS is originally Y up.
- Euler angles are used
- In the ASF file the order is given left to right so that an order of “XYZ” is: vM = vXYZ
- .amc file records the axis data of each bone frame by frame
.amc for motion
#!OML:ASF H:\Terrain\Patient Classification 1\PLAYGROUND\JustinFriday\JustinFriday.ASF
:FULLY-SPECIFIED
:DEGREES
1 // Frame numbers
root 9.37216 17.8693 -17.3198 -2.01677 -7.59696 -3.23164
lowerback 2.30193 -0.395121 1.17299
upperback 0.0030495 -0.462657 2.70388
thorax -1.27453 -0.231833 2.13151
lowerneck -9.32819 -3.76531 -6.70788
upperneck 27.8377 -3.2335 -3.01318
head 10.556 -2.55728 -0.318388
rclavicle 3.64024e-015 -6.75868e-015
rhumerus -29.5133 -11.7797 -80.4307
rradius 21.1829
rwrist -7.55893
rhand -17.4806 -21.0413
rfingers 7.12502
rthumb 8.77158 -50.8391
lclavicle 3.64024e-015 -6.75868e-015
lhumerus 17.2039 -14.515 62.7889
lradius 136.231
lwrist 10.1195
lhand -37.631 -17.4438
lfingers 7.12502
lthumb -10.6834 12.2646
rfemur -0.629535 4.65229 22.5467
rtibia 26.4457
rfoot -15.2124 -9.97437
rtoes 3.93605
lfemur 4.00236 1.20472 -13.8412
ltibia 20.088
lfoot -16.1868 6.57726
ltoes -4.61789
2
root 9.37285 17.8666 -17.3192 -2.06376 -7.58832 -3.1009
lowerback 2.29991 -0.349181 1.09181
...
...
...
Interpretation
As shown by the data hierarchy, human motions are represented as the translation offset of the root, and the rotation offsets of its child bones.
To get WCS of a specific bone segment, we must calculate the WCS of its parent, recursively to the root.
L = CinvMCB
Other useful references
http://research.cs.wisc.edu/graphics/Courses/cs-838-1999/Jeff/ASF-AMC.html
http://graphics.cs.cmu.edu/nsp/course/15-464/Fall05/assignments/StartupCodeDescription.html
http://web.cse.ohio-state.edu/~parent.1/classes/888/Mocap/index.html
http://www.darwin3d.com/gamedev/articles/col0198.pdf
http://mukai-lab.org/content/MotionCaptureDataFile.pdf
https://blog.csdn.net/zb1165048017/article/details/49358089
https://blog.csdn.net/zb1165048017/article/details/49339493
http://studentnet.cs.manchester.ac.uk/resources/library/3rd-year-projects/2016/jiayun.wang.pdf
MOCAP TOOLBOX
There are several existing projects available in Github for processing and visualization of the CMU MOCAP .asf/.amc file.
Here is one used by most researchers:
https://github.com/lawrennd/mocap
>> % Read .asf skeleton file
>> skel_86 = acclaimReadSkel('examples/86.asf');
>> % Read .amc channels file
>> [channels_86_10, skel_86] = acclaimLoadChannels('examples/86_10.amc', skel);
>> % Visualize the skeleton in motion
>> skelPlayData(skel_86, channels_86_10, 1/120);
Be aware that this matlab code was written more than a decade ago and it uses an obselete function of getline which will cause problems if run in a newer version of matlab. The solution is to switch to another function of fgetl.
Modified version: https://github.com/hanspond/mocap
Visualization of subject 02 motion 02
Visualization of subject 02 motion 02 with a ellipsoidal model

Others tool:
https://github.com/CalciferZh/AMCParser
https://github.com/CalciferZh/SMPL
https://github.com/surenkum/c3d_to_xyz
https://github.com/CalciferZh/CMU-MoCap-Washer
A list to other MOCAP databases:
http://www.jeroenvanboxtel.com/MocapDatabases.html
վ HᴗP ի
This blog is under a CC BY-NC-SA 3.0 Unported License
Link to this article: https://hanspond.github.io/2018/08/10/MOCAP data extraction and visualization/
